Research Topics
- Mechanisms of genome evolution
- Transposable elements
- Plant-microbe interactions
Genomics and bioinformatics in plants and fungi
Genomics
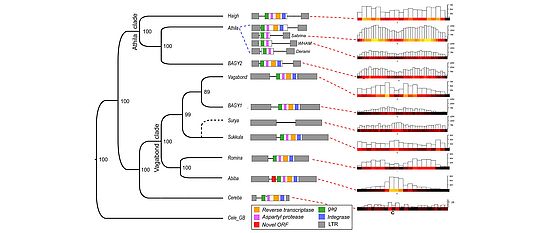
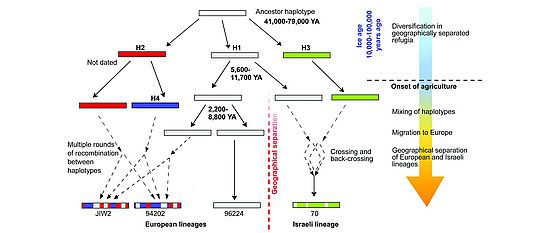
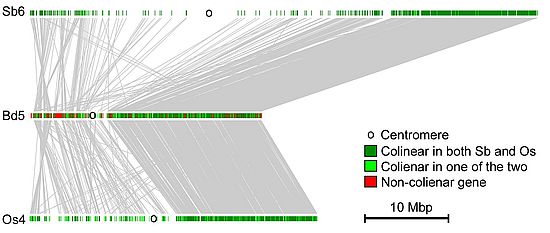
Plant and fungal genomes are very dynamic and evolve much more rapidly than animal genomes. We are investigating the molecular mechanisms that drive this rapid evolution. Genomes are expanded by the amplification of transposable elements. The expansion of genomes is counteracted by deletion of DNA. We compare gene content, gene order and repeat content of different species. Comparative analysis is a powerful method to discover mechanisms that drive the evolution of genomes. We have so discovered mechanisms that cause gene duplications, gene movement and gene loss.
Using comparative genomics and transcriptomics, we also study interactions of plants and microbes. This includes plant/pathogen interactions as well as symbioses. We were involved in sequencing and analysis of the genome of wheat powdery mildew and the study of genetic diversity of the pathogen. Additionally, we study symbiotic systems such as lichens and the symbiosis of Psychotria plants with Burkholderia. An important focus of our work is the development of bioinformatics tools that allow complex analyses on large datasets for whole-genome analysis, comparative genomics and data visualization.
Recent Publications
► Google Scholar: https://scholar.google.ch/citations?hl=en&user=BvIWQ5sAAAAJ&view_op=list_works&sortby=pubdate
- Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins (vol 7, 327, 2021)
J Sanchez-Martin, V Widrig, G Herren, T Wicker, H Zbinden, J Gronnier, ...
NATURE PLANTS, 2023, 10.1038/s41477-023-01375-3 - Comparative Transcriptome Analysis of Wheat Lines in the Field Reveals Multiple Essential Biochemical Pathways Suppressed by Obligate Pathogens
Poretti, Manuel; Sotiropoulos, Alexandros G.; Graf, Johannes; Jung, Esther; Bourras, Salim; Krattinger, Simon G.; Wicker, Thomas
Frontiers In Plant Science 10.3389/fpls.2021.720462 SEP 29 2021 - A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen
Oggenfuss, Ursula; Badet, Thomas; Wicker, Thomas; Hartmann, Fanny E.; Singh, Nikhil Kumar; Abraham, Leen; Karisto, Petteri; Vonlanthen, Tiziana; Mundt, Christopher; McDonald, Bruce A.; Croll, Daniel
Elife 10.7554/eLife.69249 SEP 16 2021 - Long-read sequence assembly: a technical evaluation in barley
Mascher, Martin; Wicker, Thomas; Jenkins, Jerry; Plott, Christopher; Lux, Thomas; et al.
Plant Cell 10.1093/plcell/koab077 JUN 2021 - Alleles of a wall-associated kinase gene account for three of the major northern corn leaf blight resistance loci in maize
Yang P; Scheuermann D; Kessel B; Koller T; Greenwood JR; Hurni S; Herren G; Zhou SH; Marande, W; Wicker T; Krattinger SG; Ouzunova M; Keller, B
Plant Journal 10.1111/tpj.15183 Published: APR 2021 - Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential
Rabanus-Wallace, M. Timothy; Hackauf, Bernd; Mascher, Martin; Lux, Thomas; Wicker, Thomas; et al.
Nature Genetics 10.1038/s41588-021-00807-0 Early Access: MAR 2021 - Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins
Sanchez-Martin, Javier; Widrig, Victoria; Herren, Gerhard; Wicker, Thomas; Zbinden, Helen; Gronnier, Julien; Sporri, Laurin; Praz, Coraline R; Heuberger, Matthias; Kolodziej, Markus C.; Isaksson, Jonatan; Steuernagel, Burkhard; Karafiatova, Miroslava; Dolezel, Jaroslav; Zipfel, Cyril; Keller, Beat
Nature Plants, 10.1038/s41477-021-00869-2 MAR 2021 - Defects in Cell Wall Differentiation of the Arabidopsis Mutant rol1-2 Is Dependent on Cyclin-Dependent Kinase CDK8
Schumacher, Isabel; Ndinyanka Fabrice, Tohnyui; Abdou, Marie-Therese; Kuhn, Benjamin M.; Voxeur, Aline; Herger, Aline; Roffler, Stefan; Bigler, Laurent; Wicker, Thomas; Ringli, Christoph
Cells 10.3390/cells10030685 Published: MAR 2021 - A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat
Kolodziej, Markus C.; Singla, Jyoti; Sanchez-Martin, Javier; Zbinden, Helen; Simkova, Hana; Karafiatova, Miroslava; Dolezel, Jaroslav; Gronnier, Julien; Poretti, Manuel; Glauser, Gaetan; Zhu, Wangsheng; Koster, Philipp; Zipfel, Cyril; Wicker, Thomas; Krattinger, Simon; Keller, Beat
Nature Communications, DOI: 10.1038/s41467-020-20777-x FEB 11 2021 - De Novo Genome Assembly of the Japanese Wheat Cultivar Norin 61 Highlights Functional Variation in Flowering Time and Fusarium-Resistant Genes in East Asian Genotypes
Shimizu, Kentaro K.; Copetti, Dario; Okada, Moeko; Wicker, Thomas; Tameshige, Toshiaki; et al.
Plant And Cell Physiology, 10.1093/pcp/pcaa152, JAN 2021 - …

Prof. Dr. Thomas Wicker
University of Zurich
Department of Plant and Microbial Biology
8008 Zurich
Tel: +41 44 63 48252
Links
Interdisciplinarity
- Collaboration with research groups in microbiology
- Collaboration with members of the Dept of Systematic and Evolutionary Botany