Evolutionary genomics of plant-pathogen interactions
Evolutionary Genetics
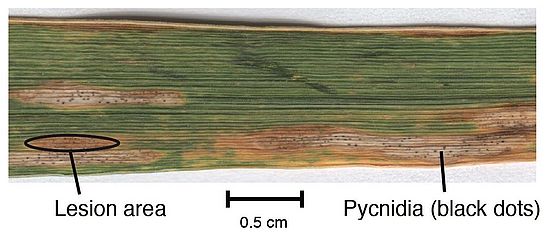
Most plants face attacks by fungal pathogens. In agriculture, outbreaks of diseases are frequent and pose a significant threat to sustainable food production. What enables pathogens to overcome host defenses and cause damage is poorly understood. A key evolutionary step for pathogens is to evolve effectors that specifically target and disable the plant immune system.
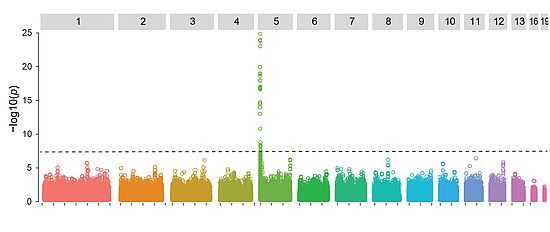
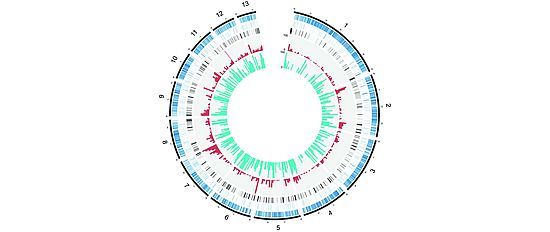
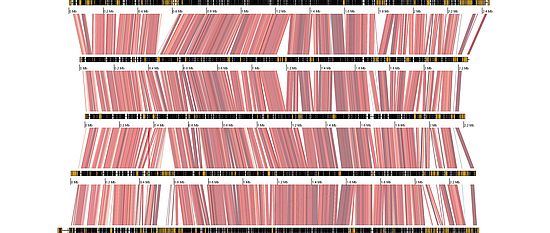
Our group uses experimental and population genomics tools to analyze the rapid emergence of pathogens. A major focus is to use genome-wide association studies (GWAS) to directly identify the genes in pathogen genomes linked to the breakdown of host resistance. We also assemble complete genomes to analyze mechanisms that generate high degrees of sequence polymorphism. We are particularly interested in the action of transposable elements (selfish elements in the genome) and the evolution of gene regulation. Our group also conducts population genomic studies to analyze the process of adaptive evolution in natural field populations.
Recent Publications
- Screening of grapevine red blotch virus in two European ampelographic collections
Reynard, Jean-Sebastien; Brodard, Justine; Dubuis, Nathalie; Kellenberger, Isabelle; Spilmont, Anne-Sophie; et al.
Journal Of Plant Pathology 10.1007/s42161-021-00952-9 NOV 2021 - Population-level deep sequencing reveals the interplay of clonal and sexual reproduction in the fungal wheat pathogen Zymoseptoria tritici
Singh, Nikhil Kumar; Karisto, Petteri; Croll, Daniel
Microbial Genomics 10.1099/mgen.0.000678 OCT 2021 - A robust sequencing assay of a thousand amplicons for the high-throughput population monitoring of Alpine ibex immunogenetics
Kessler, Camille; Brambilla, Alice; Waldvogel, Dominique; Camenisch, Glauco; Biebach, Iris; et al.
Molecular Ecology Resources10.1111/1755-0998.13452 JAN 2022 - Transcriptome-wide SNPs for Botrychium lunaria ferns enable fine-grained analysis of ploidy and population structure
Mossion, Vinciane; Dauphin, Benjamin; Grant, Jason; Kessler, Michael; Zemp, Niklaus; Croll, Daniel
Molecular Ecology Resources 10.1111/1755-0998.13478 JAN 2022 - A new method to determine the diet of pygmy hippopotamus in Tai National Park, Cote d'Ivoire
Hendier, Alba; Chatelain, Cyrille; Du Pasquier, Pierre-Emmanuel; Paris, Monique; Ouattara, Karim; et al.
African Journal Of Ecology 10.1111/aje.12888 DEC 2021 - Soil composition and plant genotype determine benzoxazinoid-mediated plant-soil feedbacks in cereals
Cadot, Selma; Gfeller, Valentin; Hu, Lingfei; Singh, Nikhil; Sanchez-Vallet, Andrea; Glauser, Gaetan; Croll, Daniel; Erb, Matthias; van der Heijden, Marcel G. A.; Schlaeppi, Klaus
Plant Cell And Environment 10.1111/pce.14184 DEC 2021 - Cryptic genetic structure and copy-number variation in the ubiquitous forest symbiotic fungus Cenococcum geophilum
Dauphin, Benjamin; Pereira, Maira de Freitas; Kohler, Annegret; Grigoriev, Igor, V; Barry, Kerrie; et al.
Environmental Microbiology 10.1111/1462-2920.15752 NOV 2021 - A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen
Oggenfuss, Ursula; Badet, Thomas; Wicker, Thomas; Hartmann, Fanny E.; Singh, Nikhil Kumar; Abraham, Leen; Karisto, Petteri; Vonlanthen, Tiziana; Mundt, Christopher; McDonald, Bruce A.; Croll, Daniel
Elife 10.7554/eLife.69249 SEP 16 2021 - Genome-wide association study for septoria tritici blotch resistance reveals the occurrence and distribution of Stb6 in a historic Swiss landrace collection (vol 217, 108, 2021)
Dutta, Anik; Croll, Daniel; McDonald, Bruce A.; Krattinger, Simon G.
Euphytica 10.1007/s10681-021-02906-9 SEP 2021 - Population genomics of transposable element activation in the highly repressive genome of an agricultural pathogen
Pereira, Danilo; Oggenfuss, Ursula; McDonald, Bruce A.; Croll, Daniel
Microbial Genomics 10.1099/mgen.0.000540 AUG 2021 - Genome-scale phylogenies reveal relationships among Parastagonospora species infecting domesticated and wild grasses
Croll, D.; Crous, P. W.; Pereira, D.; Mordecai, E. A.; McDonald, B. A.; Brunner, P. C.
Persoonia 10.3767/persoonia.2021.46.04 JUN 2021 - Machine-learning predicts genomic determinants of meiosis-driven structural variation in a eukaryotic pathogen
Badet, Thomas; Fouche, Simone; Hartmann, Fanny E.; Zala, Marcello; Croll, Daniel
Nature Communications 10.1038/s41467-021-23862-x JUN 10 2021 - Rapid sequence evolution driven by transposable elements at a virulence locus in a fungal wheat pathogen
Singh, Nikhil Kumar; Badet, Thomas; Abraham, Leen; Croll, Daniel
Bmc Genomics10.1186/s12864-021-07691-2 MAY 27 2021 - Mapping the adaptive landscape of a major agricultural pathogen reveals evolutionary constraints across heterogeneous environments
Dutta, Anik; Hartmann, Fanny E.; Francisco, Carolina Sardinha; McDonald, Bruce A.; Croll, Daniel
Isme Journal, 10.1038/s41396-020-00859-w MAY 2021 - Emergence and diversification of a highly invasive chestnut pathogen lineage across southeastern Europe
Stauber, Lea; Badet, Thomas; Feurtey, Alice; Prospero, Simone; Croll, Daniel
Elife, 10.7554/eLife.56279 MAR 5 2021 - …
Publications 2020
- The Genetic Architecture of Emerging Fungicide Resistance in Populations of a Global Wheat Pathogen
Pereira, Danilo; McDonald, Bruce A.; Croll, Daniel
Genome Biology And Evolution, 10.1093/gbe/evaa203 DEC 2020 - The complex genomic basis of rapid convergent adaptation to pesticides across continents in a fungal plant pathogen
Hartmann, Fanny E.; Vonlanthen, Tiziana; Singh, Nikhil Kumar; McDonald, Megan C.; Milgate, Andrew; et al.
Molecular Ecology, 10.1111/mec.15737 DEC 2020 - Tackling microbial threats in agriculture with integrative imaging and computational approaches
Singh, Nikhil Kumar; Dutta, Anik; Puccetti, Guido; Croll, Daniel
Computational And Structural Biotechnology Journal 10.1016/j.csbj.2020.12.018 2020 - A Chromosome-Scale Genome Assembly for the Fusarium oxysporum Strain Fo5176 To Establish a Model Arabidopsis-Fungal Pathosystem
Fokkens, Like; Guo, Li; Dora, Susanne; Wang, Bo; Ye, Kai; Sanchez-Rodriguez, Clara; Croll, Daniel
G3-genes Genomes Genetics DOI: 10.1534/g3.120.401375 OCT 2020 - Transcriptome plasticity underlying plant root colonization and insect invasion by Pseudomonas protegens
Vesga, Pilar; Flury, Pascale; Vacheron, Jordan; Keel, Christoph; Croll, Daniel; Maurhofer M
Isme Journal DOI: 10.1038/s41396-020-0729-9 NOV 2020 - A Chromosome-Scale Genome Assembly for the Fusarium oxysporum Strain Fo5176 To Establish a Model Arabidopsis-Fungal Pathosystem
Fokkens, Like; Guo, Li; Dora, Susanne; Wang, Bo; Ye, Kai; Sanchez-Rodriguez, Clara; Croll, Daniel
G3-genes Genomes Genetics 10.1534/g3.120.401375 OCT 2020 - Comparative Genomics Analyses of Lifestyle Transitions at the Origin of an Invasive Fungal Pathogen in the Genus Cryphonectria
Stauber, Lea; Prospero, Simone; Croll, Daniel
Msphere DOI: 10.1128/mSphere.00737-20 SEP-OCT 2020 - Maintenance of variation in virulence and reproduction in populations of an agricultural plant pathogen
Dutta, Anik; Croll, Daniel; McDonald, Bruce A.; Barrett, Luke G.
Evolutionary Applications DOI: 10.1111/eva.13117 SEP 2020 - The rise and fall of genes: origins and functions of plant pathogen pangenomes
Badet, Thomas; Croll, Daniel
Current Opinion In Plant Biology DOI: 10.1016/j.pbi.2020.04.009 AUG 2020 - Genome compartmentalization predates species divergence in the plant pathogen genus Zymoseptoria
Feurtey, Alice; Lorrain, Cecile; Croll, Daniel; Eschenbrenner, Christoph; Freitag, Michael; et al.
Bmc Genomics DOI: 10.1186/s12864-020-06871-w AUG 26 2020 - Natural selection drives population divergence for local adaptation in a wheat pathogen
Pereira, Danilo; Croll, Daniel; Brunner, Patrick C.; McDonald, Bruce A.
Fungal Genetics And Biology DOI: 10.1016/j.fgb.2020.103398 AUG 2020 - A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization
Sanchez-Vallet, Andrea; Tian, Hui; Rodriguez-Moreno, Luis; Valkenburg, Dirk-Jan; Saleem-Batcha, Raspudin; et al.
Plos Pathogens DOI: 10.1371/journal.ppat.1008652 JUN 2020 - Chromosomal assembly and analyses of genome-wide recombination rates in the forest pathogenic fungus Armillaria ostoyae
Heinzelmann, Renate; Rigling, Daniel; Sipos, Gyorgy; Muensterkoetter, Martin; Croll, Daniel
Heredity DOI: 10.1038/s41437-020-0306-z MAR 2020 - Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex
Grossen, Christine; Guillaume, Frederic; Keller, Lukas F.; Croll, Daniel
Nature Communications, 11(1) Number: 1001 DOI: 10.1038/s41467-020-14803-1 FEB 21 2020 - A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici
Badet, Thomas; Oggenfuss, Ursula; Abraham, Leen; McDonald, Bruce A.; Croll, Daniel
Bmc Biology 18(1) Article Number: 12 DOI: 10.1186/s12915-020-0744-3 FEB 11 2020 - Stress-Driven Transposable Element De-repression Dynamics and Virulence Evolution in a Fungal Pathogen
Fouche, Simone; Badet, Thomas; Oggenfuss, Ursula; Plissonneau, Clemence; Francisco, Carolina Sardinha; Croll Daniel
Molecular Biology And Evolution DOI: 10.1093/molbev/msz216 JAN 2020 - …

Prof. Dr. Daniel Croll
University of Neuchâtel
Institute of Biology
2000 Neuchâtel
Tel: +41 (0)32 718 23 30
Research topics
- Genetic basis of pathogen virulence
- Ecological genomics and local adaptation of pathogens
- Population genomics of pathogen outbreaks
- Structure and evolution of pathogen genomes
- Transposable elements and the evolution of gene regulation
Interdisciplinary
- Crop resistance breeding
- Bioinformatics and genome assembly
- Molecular plant pathology
- Fungal community analyses
- Genome-wide association mapping (GWAS)