Chromatin and transcriptome dynamics during sexual plant reproduction
Molecular Genetics of Plant Development
Sexual reproduction in flowering plants involves several cell fate transitions: (i) vegetative-to-reproductive transition of the apical meristem, (ii) reprogramming of floral somatic cells to generate the gametic progenitors (micro/mega-spore mother cells), (iii) differentiation of specialized gametes, (iv) fertilization and development of a totipotent embryo. These transitions occurring in the adult plant illustrate the remarkable plasticity of plant cells, unlike animals’ which are committed early during embryogenesis. My research focuses on transitions and cell fate reprogramming in the transitions (ii-iv) and investigates two critical aspects:
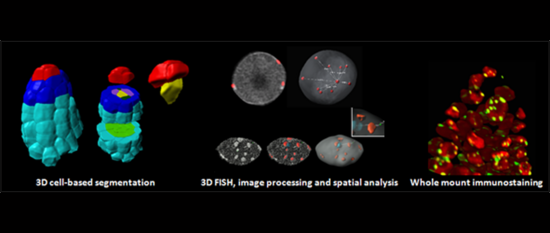
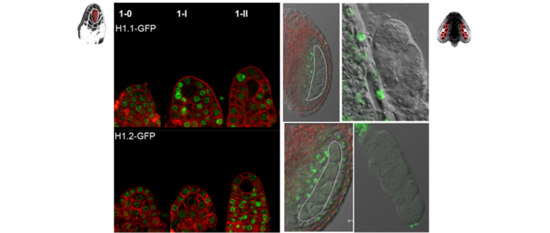
(1) From the diploid gamete progenitor to the differentiated gametes the plant chromatin seem to undergo dramatic and dynamic changes not only in histone/DNA modifications but also in structural organisation (fig1) and in core nucleosome constituent, as seen in chromatin staining and GFP reporter studies. We are investigating these key processes underlying fate reprogramming.
(2) fertilization unites two parental genomes to form a totipotent embryo; yet parental alleles do not contribute in an equal way to the embryonic transcriptome which remains massively maternally dominant, as seen in allele-specific profiling experiments (fig 2).
Recent Publications
- Three-dimensional genome organization in epigenetic regulations: cause or consequence?
Baroux, Celia
Current Opinion In Plant Biology 10.1016/j.pbi.2021.102031 JUN 2021 - Organ geometry channels reproductive cell fate in the Arabidopsis ovule primordium
Hernandez-Lagana, Elvira; Mosca, Gabriella; Mendocilla-Sato, Ethel; Pires, Nuno; Frey, Anja; et al.
Elife 10.7554/eLife.66031 MAY 7 2021 - Impact of Transposable Elements on Methylation and Gene Expression across Natural Accessions of Brachypodium distachyon
Wyler, Michele; Stritt, Christoph; Walser, Jean-Claude; Baroux, Celia; Roulin, Anne C.
Genome Biology And Evolution, 10.1093/gbe/evaa180 NOV 2020 - Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis
Rutowicz, K; Lirski, M; Mermaz, B; Teano, G; Schubert, J; Mestiri, I; Kroten, MA; Fabrice, TN; Fritz, S; Grob, S; Ringli, C; Cherkezyan, L; Barneche, F; Jerzmanowski, A; Baroux, C
GENOME BIOLOGY, 20 (1):10.1186/s13059-019-1767-3 AUG 7 2019 - The SMC5/6 Complex Subunit NSE4A Is Involved in DNA Damage Repair and Seed Development
Diaz, M; Pecinkova, P; Nowicka, A; Baroux, C; Sakamoto, T; Gandha, PY; Jerabkova, H; Matsunaga, S; Grossniklaus, U; Pecinka, A
PLANT CELL, 31 (7):1579-1597; 10.1105/tpc.18.00043 JUL 2019 - Probing the 3D architecture of the plant nucleus with microscopy approaches: challenges and solutions
Dumur, T; Duncan, S; Graumann, K; Desset, S; Randall, RS; Scheid, OM; Prodanov, D; Tatout, C; Baroux, C
NUCLEUS, 10 (1):181-212; 10.1080/19491034.2019.1644592 JAN 1 2019 - Seeds – An evolutionary innovation underlying reproductive success in flowering plants
Baroux, C; Grossniklaus, U
PLANT DEVELOPMENT AND EVOLUTION, 131 605-+; 10.1016/bs.ctdb.2018.11.017 2019
Current Topics in Developmental Biology - Meeting report - INDEPTH kick-off meeting
Parry, G; Probst, AV; Baroux, C; Tatout, C
JOURNAL OF CELL SCIENCE, 131 (12):10.1242/jcs.220558 JUN 2018 - Aberrant imprinting may underlie evolution of parthenogenesis
Kirioukhova, O; Shah, JN; Larsen, DS; Tayyab, M; Mueller, NE; Govind, G; Baroux, C; Federer, M; Gheyselinck, J; Barrell, PJ; Ma, H; Sprunck, S; Huettel, B; Wallace, H; Grossniklaus, U; Johnston, AJ
SCIENTIFIC REPORTS, 8 https://doi.org/10.1038/s41598-018-27863-7 JUL 13 2018 - …

PD Dr. Célia Baroux
University of Zurich
Department of Plant and Microbial Biology
8008 Zurich
Tel: +41 (0)44 634 82 50
Research topics
- Chromatin dynamics during female gametogenesis reprogramming
- Dynamic parental contributions to the early embryonic transcriptome