3D-omics
Epigenetics
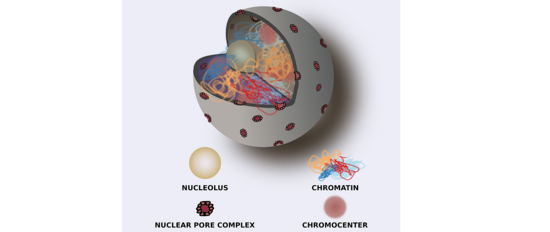
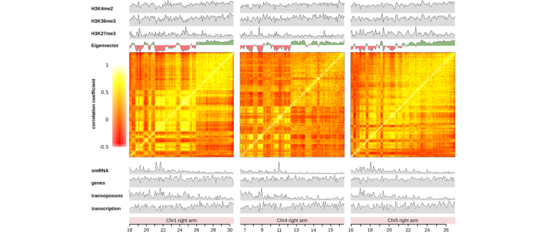
We work on the functional roles of three-dimensional (3D) chromosomal architecture. To show that 3D-chromosome folding is much more than packing of long chromosomes into tiny nuclei, we are using both, state-of-the-art molecular techniques such as Hi-C and classic genetics approaches.
Our interest focuses on the interplay of the different genome organization levels, especially the relationship between the 3D-genome and the two other organizational levels, the DNA sequence and the epigenome. We are intrigued by the possibility that 3D-chromatin folding may contain information and, hence, may exert an important biological function in both, micro-scale cellular mechanisms and, macro-scale evolution.
As for micro-scale processes, a specific 3D-chromatin structure, termed the KNOT is involved in the genome’s immune system. The KNOT is formed by tight 3D contacts among ten KNOT Entangled Elements (KEEs), which are enriched in both transposable elements and associated small RNAs. Our current research aims at investigating the underlying mechanisms required to form the KNOT. Thereby, we expect to reveal important factors regulating 3D-genome architecture.
Recent Publications
- Two-step regulation of centromere distribution by condensin II and the nuclear envelope proteins
Sakamoto, Takuya; Sakamoto, Yuki; Grob, Stefan; Slane, Daniel; Yamashita, Tomoe; et al.
Nature Plants 10.1038/s41477-022-01200-3 AUG 2022 - Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies (vol 5, 50, 2018)
Zhang, Lei; Cai, Xu; Wu, Jian; Liu, Min; Grob, Stefan; Cheng, F; Liang, JL; Cai, CC; Liu, ZY; Liu, B; Wang, F; Li, S; Liu, FY; Li, XM; Cheng, L; Yang, WC; Li, MH; Grossniklaus, U; Zheng, HK; Wang, XW
Horticulture Research, DOI: 10.1038/s41438-019-0210-y - Haplotype-resolved genomes of geminivirus-resistant and geminivirus-susceptible African cassava cultivars
Kuon, JE; Qi, WH; Schlapfer, P; Hirsch-Hoffmann, M; von Bieberstein, PR; Patrignani, A; Poveda, L; Grob, S; Keller, M; Shimizu-Inatsugi, R; Grossniklaus, U; Vanderschuren, H; Gruissem, W
BMC BIOLOGY, 17 (1):10.1186/s12915-019-0697-6 SEP 18 2019 - Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis
Rutowicz, K; Lirski, M; Mermaz, B; Teano, G; Schubert, J; Mestiri, I; Kroten, MA; Fabrice, TN; Fritz, S; Grob, S; Ringli, C; Cherkezyan, L; Barneche, F; Jerzmanowski, A; Baroux, C
GENOME BIOLOGY, 20 (1):10.1186/s13059-019-1767-3 AUG 7 2019 - Invasive DNA elements modify the nuclear architecture of their insertion site by KNOT-linked silencing in Arabidopsis thaliana
Grob, S; Grossniklaus, U
GENOME BIOLOGY, 20 10.1186/s13059-019-1722-3 JUN 11 2019 - Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies
Zhang, L; Cai, X; Wu, J; Liu, M; Grob, S; Cheng, F; Liang, JL; Cai, CC; Liu, ZY; Liu, B; Wang, F; Li, S; Liu, FY; Li, XM; Cheng, L; Yang, WC; Li, MH; Grossniklaus, U; Zheng, HK; Wang, XWHORTICULTURE RESEARCH, 5 10.1038/s41438-018-0071-9 AUG 15 2018 - Technical Review: A Hitchhiker's Guide to Chromosome Conformation Capture.
Grob S, Cavalli G.
Methods Mol Biol. 2018;1675:233-246. 10.1007/978-1-4939-7318-7_14. - Plants are not so different
Grob, Stefan
Nat Plants. 2017 Sep;3(9):690-691. 10.1038/s41477-017-0013-9. - Circular Chromosome Conformation Capture in Plants.
Grob S.
Methods Mol Biol. 2017;1610:73-92. doi: 10.1007/978-1-4939-7003-2_6. - Chromosome conformation capture-based studies reveal novel features of plant nuclear architecture.
Grob S, Grossniklaus U.
Curr Opin Plant Biol. 36:149-157. doi: 10.1016/j.pbi.2017.03.004. Epub 2017 Apr 12. Review. - Chromatin Conformation Capture-Based Analysis of Nuclear Architecture.
Grob S, Grossniklaus U.
Methods Mol Biol. 2017;1456:15-32. - HiCdat: a fast and easy-to-use Hi-C data analysis tool.
Schmid MW, Grob S, Grossniklaus U.
BMC Bioinformatics. 2015 Sep 3;16:277. doi: 10.1186/s12859-015-0678-x.

Dr. Stefan Grob
University of Zurich
Department of Plant and Microbial Biology
8008 Zurich
Tel: +41 44 644 8250
Research topics
- Epigenetics
- Gene silencing
- Defense against parasitic DNA elements
- 3D chromosome architecture
- Genome evolution
Interdisciplinary
- Genome evolution
- Genome assemblies
- Analysis of -omics data
- Transcriptional regulation by 3D contacts, such as promoter-enhancer contacts